Research
Deep-profiling tissue-resident memory T cells during viral infection and virus-associated cancer
Current knowledge of human T cell biology is often limited to the study of total T cell population without the understanding of antigen-specific immunity due to the lack of apparent T cell epitopes. In addition, most of the studies only investigate peripheral T cells without encompassing site-specific responses in human tissues, where the diseases usually occur. These prevent the accurate interpretation of disease-specific immune responses and restrain the therapeutic implications of findings. Our goal is to dissect the complexity of virus-specific CD8+ T cell responses detected in healthy versus diseased blood and tissue samples. We are particularly interested in chronic viral (e.g., hepatitis B virus, HBV) infection, HBV-associated liver diseases, and SARS-CoV-2 infection. Combining the usages of highly multiplexed combinatorial peptide-MHC tetramer staining, mass cytometry, and other single-cell multi-omics, we aim to comprehensively investigate the T cell antigen-specificities, virus-reactive T cell receptors (TCR), and cellular phenotypes across human tissues, with a focus on the characteristics of tissue-resident memory T (TRM) cells and exhausted T (TEX) cells. We believe our work can facilitate the fundamental understanding of human T cell biology, improve immunotherapeutics against cancer, and advance the next generation of vaccine design.
JRNLclub talk
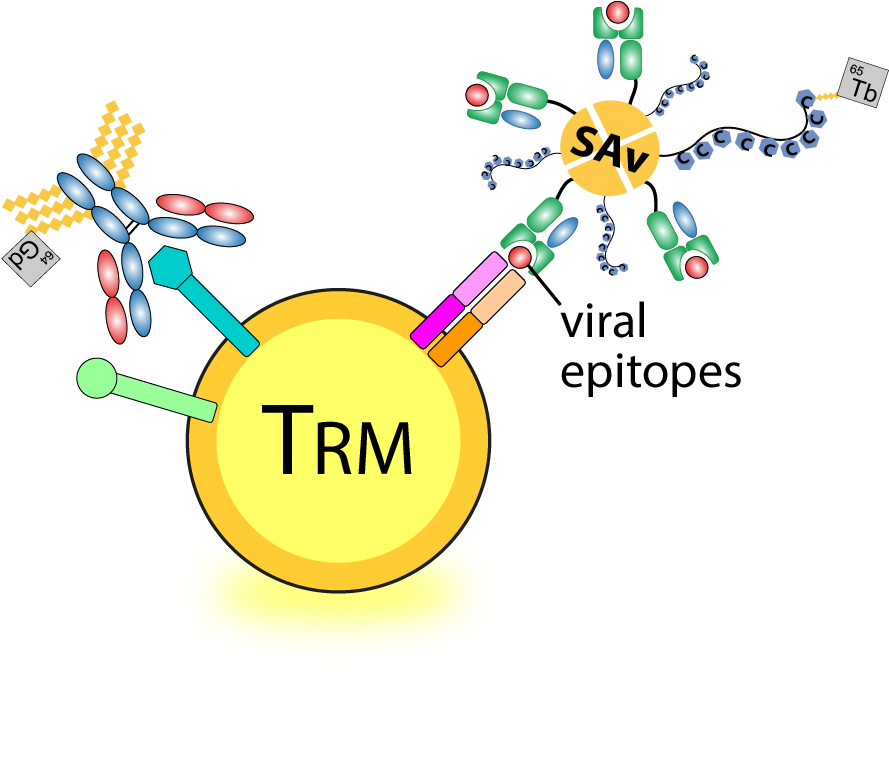
Defining long-term immune protection of human airway tissue-resident memory SARS-CoV-2-specific T cells against breakthrough infection
Our team aims to comprehensively quantify and qualify SARS-CoV-2-specific T cells against mutated and conserved viral epitopes across various human respiratory tract tissues in human. We focus on the T cells immunity specific for common HLA alleles in Asia Pacific, and their cross-reactivity against SARS-CoV-2 VOCs, and further characterize their phenotypic and transcriptomic profiles of these airway SARS-CoV-2-specific TRM cells. Our long-term goal is to define the immunological memory of SARS-CoV-2-specific T cells that provide broad immune protection against VOCs-induced breakthrough infection and the longevity of the memory T cell responses in post COVID-19 era.
Investigating the functional profiles of HBV-specific adaptive immunity in chronic HBV and HBV-associated liver diseases
One of the central research theme in our laboratory is to dissect the HBV-specific T and B cell responses in great details, with the focus on the immune populations reside in human liver. We leverage the highly multiplexed combinatorial peptide-MHC tetramer strategy and mass cytometry to identify the relevant HBV-specific T cells isolated from clinical samples in various types of diseases, including patients with chronic HBV infection, HBV-associated HCC, and HBV-associated liver failure, as well as MASH patients at the background of HBV infection. We use scRNA-seq, scTCR-seq, scATAC-seq, single-cell spatial transcriptome to further characterize their specificity, quantity, and quality of the disease relevant cells.
Using multimodal machine learning to visualize in situ intrahepatic HBV-specific T cells in human liver
In conjunction with various approaches, we are building a machine learning framework to visualize the localization of HBV-specific TRM cells and study their spatial dynamics in native human liver tissues derived from patients with HBV-associated liver diseases.
We are collaborating with Singapore General Hospital / National Cancer Center Singapore, Garvan Institute of Medical Research in Australia, and ASTRA consortium for this project.
Asia-Pacific Spatial Translational Research Alliance (ASTRA) consortium
Our laboratory is one of the core member of ASTRA consortium (initiated by The Garvan Institute of Medical Research and University of Tokyo) that aims to build a +2000 pan cancer spatial transcriptome and proteome atlas in Asia-Pacific Region, including Japan, Australia, India, Taiwan, Korea, Singapore, Thailand, Hong Kong, and China. Through this joint force, we aim to decode the spatial dynamics of virus-specific T cell responses and the immune landscape of tertiary lymphoid structure in tumor microenvironment. We are particularly focusing on liver diseases.

Using cutting-edge methods to explore and visualize the high-dimensional complexity of immune cells
The lab is experienced in combining the utility of mass cytometry (CyTOF), high-parameter spectral flow cytometry, single-cell analytical tools, and high-dimensional analysis, to delve into the biological insights of human immune responses.